Scattering Function Normalization¶
Scott Prahl
July 2022
[1]:
#!pip install --user miepython
[2]:
import numpy as np
import matplotlib.pyplot as plt
try:
import miepython
except ModuleNotFoundError:
print('miepython not installed. To install, uncomment and run the cell above.')
print('Once installation is successful, rerun this cell again.')
Solid Angles¶
Solid angles are the 3D analog of 2D angles. A radian \(\theta\) is defined as the angle represented by an arc length on circle divided by the radius \(R\) of the circle
A steradian \(\Omega\) is defined as the area on a surface of a sphere divided by the square of its radius \(R\).
Thus the total number of steradians in a sphere is \(4\pi R^2/R^2=4\pi\)
For example usage, a circular detector (pointed at the center) with radius \(r_d\) at a distance \(R\) from the center will subtend an approximate angle (the detector is flat and does not curve with the sphere surface)
(assuming \(r_d\ll R\)). Now if \(P_0\) of light is scattered by a sphere located at the center then the scattered power on the detector will be
where \(\mu\) is the cosine of the angle between the incoming light and a ray to the center of the detector.
Normalization of the scattered light¶
Mie scattering is used in a wide variety of disciplines to describe the scattering pattern from spheres. Not surprisingly a bunch of different normalizations have arisen that can be described based on the integrating the scattering function over all directions (\(4\pi\) steradians).
Integrating the scattering phase function all solid angles suggests the following dimensionless possibilities which have been chosen by one or more authors for their convenience. miepython now supports all of these.
where \(x=2\pi a/\lambda\) is the size parameter of a sphere with radius \(a\) at wavelength \(\lambda\).
The single scattering albedo is \(a\), defined as
and \(Q_\mathrm{sca}\), \(Q_\mathrm{ext}\), and \(Q_\mathrm{abs}\) are the scattering, extinction, and absorption efficiencies.
Verifying normalization¶
Since \(d\Omega=\sin\theta d\theta\,d\phi\) is a differential solid angle, the integral of the scattering phase function over all solid angles is
or with a change of variables \(\mu=\cos\theta\) and using symmetry (the phase function is independent of \(\phi\)), the integral simplifies to
This integral can be done numerically by simply summing all the rectangles
which can be found using np.trapz()
[3]:
## Phase Functions
Case 1. n=1.5, x=1¶
For this non-strongly peaked scattering function, the simple integration remains close to the expected value.
[4]:
m = 1.5
x = 1
mu = np.linspace(-1,1,501)
intensity = miepython.i_unpolarized(m,x,mu)
qext, qsca, qback, g = miepython.mie(m,x)
norms = ['albedo','one','4pi','qsca','qext','bohren','wiscombe']
expected = [qsca/qext,1.0,4*np.pi,qsca,qext,4*np.pi*x**2*qsca,np.pi*x**2*qsca]
plt.plot(mu,intensity)
plt.xlabel(r'$\cos(\theta)$')
plt.ylabel('Unpolarized Scattering Intensity [1/sr]')
plt.title('m=%.3f%+.3fj, x=%.2f, default normalization'%(m.real,m.imag,x))
plt.show()
print(' Normalization Total Expected')
for i,norm in enumerate(norms):
intensity = miepython.i_unpolarized(m,x,mu,norm)
total = 2 * np.pi * np.trapz(intensity, mu)
print("%14s %8.3f %8.3f" % (norm, total, expected[i]))
Normalization Total Expected
albedo 1.000 1.000
one 1.000 1.000
4pi 12.566 12.566
qsca 0.215 0.215
qext 0.215 0.215
bohren 2.703 2.703
wiscombe 0.676 0.676
Case 2: m=1.5-1.5j, x=1¶
For this non-strongly peaked scattering function, the simple integration remains close to the expected value.
[5]:
m = 1.5 - 1.5j
x = 1
mu = np.linspace(-1,1,501)
intensity = miepython.i_unpolarized(m,x,mu)
qext, qsca, qback, g = miepython.mie(m,x)
norms = ['albedo','one','4pi','qsca','qext','bohren','wiscombe']
expected = [qsca/qext,1.0,4*np.pi,qsca,qext,4*np.pi*x**2*qsca,np.pi*x**2*qsca]
plt.plot(mu,intensity)
plt.xlabel(r'$\cos(\theta)$')
plt.ylabel('Unpolarized Scattering Intensity [1/sr]')
plt.title('m=%.3f%+.3fj, x=%.2f, default normalization'%(m.real,m.imag,x))
plt.show()
print(' Normalization Total Expected')
for i,norm in enumerate(norms):
intensity = miepython.i_unpolarized(m,x,mu,norm)
total = 2 * np.pi * np.trapz(intensity, mu)
print("%14s %8.3f %8.3f" % (norm, total, expected[i]))
Normalization Total Expected
albedo 0.377 0.377
one 1.000 1.000
4pi 12.566 12.566
qsca 1.139 1.139
qext 3.023 3.023
bohren 14.308 14.308
wiscombe 3.577 3.577
Case III, evenly spaced \(\theta\)¶
For this non-strongly peaked scattering function, even spacing in \(\theta\) improves the accuracy of the integration.
[6]:
m = 1.5-1.5j
x = 2
theta = np.linspace(np.pi,0,361)
mu = np.cos(theta)
intensity = miepython.i_unpolarized(m,x,mu)
qext, qsca, qback, g = miepython.mie(m,x)
norms = ['albedo','one','4pi','qsca','qext','bohren','wiscombe']
expected = [qsca/qext,1.0,4*np.pi,qsca,qext,4*np.pi*x**2*qsca,np.pi*x**2*qsca]
plt.plot(mu,intensity)
plt.xlabel(r'$\cos(\theta)$')
plt.ylabel('Unpolarized Scattering Intensity [1/sr]')
plt.title('m=%.3f%+.3fj, x=%.2f, default normalization'%(m.real,m.imag,x))
plt.show()
print(' Normalization Total Expected')
for i,norm in enumerate(norms):
intensity = miepython.i_unpolarized(m,x,mu,norm)
total = 2 * np.pi * np.trapz(intensity, mu)
print("%14s %8.3f %8.3f" % (norm, total, expected[i]))
Normalization Total Expected
albedo 0.496 0.496
one 1.000 1.000
4pi 12.567 12.566
qsca 1.523 1.523
qext 3.069 3.068
bohren 76.579 76.576
wiscombe 19.145 19.144
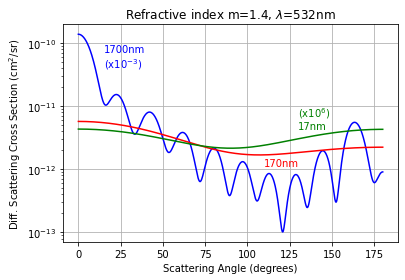
Differential Scattering Cross Section¶
The differential scattering cross section \(\frac{d\sigma_{sca}}{d\Omega}\) is defined in terms of the total scattering cross section
Thus if the unpolarized scattering is normalized so its integral is the scattering efficiency
then
The differential scattering cross section can be obtained miepython by normalizing to qsca and multiplying the result by the geometric cross section
diff_sca = np.pi * r**2 * miepython.i_unpolarized(m,x,mu,norm='qsca')
For example, here is a replica of figure 4
[7]:
m = 1.4-0j
lambda0 = 532e-9 # m
theta = np.linspace(0,180,1000)
mu = np.cos(theta* np.pi/180)
d = 1700e-9 # m
x = 2 * np.pi/lambda0 * d/2
geometric_cross_section = np.pi * d**2/4 * 1e4 # cm**2
qext, qsca, qback, g = miepython.mie(m,x)
sigma_sca = geometric_cross_section * miepython.i_unpolarized(m,x,mu,'qsca')
plt.semilogy(theta, sigma_sca*1e-3, color='blue')
plt.text(15, sigma_sca[0]*3e-4, "%.0fnm\n(x10$^{-3}$)" % (d*1e9), color='blue')
d = 170e-9 # m
x = 2 * np.pi/lambda0 * d/2
geometric_cross_section = np.pi * d**2/4 * 1e4 # cm**2
qext, qsca, qback, g = miepython.mie(m,x)
sigma_sca = geometric_cross_section * miepython.i_unpolarized(m,x,mu,'qsca')
plt.semilogy(theta, sigma_sca, color='red')
plt.text(110, sigma_sca[-1]/2, "%.0fnm" % (d*1e9), color='red')
d = 17e-9 # m
x = 2 * np.pi/lambda0 * d/2
geometric_cross_section = np.pi * d**2/4 * 1e4 # cm**2
qext, qsca, qback, g = miepython.mie(m,x)
sigma_sca = geometric_cross_section * miepython.i_unpolarized(m,x,mu,'qsca')
plt.semilogy(theta, sigma_sca*1e6, color='green')
plt.text(130, sigma_sca[-1]*1e6, "(x10$^6$)\n%.0fnm" % (d*1e9), color='green')
plt.title("Refractive index m=1.4, $\lambda$=532nm")
plt.xlabel("Scattering Angle (degrees)")
plt.ylabel("Diff. Scattering Cross Section (cm$^2$/sr)")
plt.grid(True)
plt.show()
Comparison to Wiscombe’s Mie Program¶
Wiscombe normalizes as
where \(p(\theta)\) is the scattered light.
Once corrected for differences in phase function normalization, Wiscombe’s test cases match those from miepython exactly.
Wiscombe’s Test Case 14¶
[8]:
"""
MIEV0 Test Case 14: Refractive index: real 1.500 imag -1.000E+00, Mie size parameter = 1.000
Angle Cosine S-sub-1 S-sub-2 Intensity Deg of Polzn
0.00 1.000000 5.84080E-01 1.90515E-01 5.84080E-01 1.90515E-01 3.77446E-01 0.0000
30.00 0.866025 5.65702E-01 1.87200E-01 5.00161E-01 1.45611E-01 3.13213E-01 -0.1336
60.00 0.500000 5.17525E-01 1.78443E-01 2.87964E-01 4.10540E-02 1.92141E-01 -0.5597
90.00 0.000000 4.56340E-01 1.67167E-01 3.62285E-02 -6.18265E-02 1.20663E-01 -0.9574
"""
x=1.0
m=1.5-1.0j
mu=np.cos(np.linspace(0,90,4) * np.pi/180)
qext, qsca, qback, g = miepython.mie(m,x)
albedo = qsca/qext
unpolar = miepython.i_unpolarized(m, x, mu, 'wiscombe')
unpolar_miev = np.array([3.77446E-01,3.13213E-01,1.92141E-01,1.20663E-01])
ratio = unpolar_miev/unpolar
print("MIEV0 Test Case 14: m=1.500-1.000j, Mie size parameter = 1.000")
print()
print(" %9.1f°%9.1f°%9.1f°%9.1f°"%(0,30,60,90))
print("MIEV0 %9.5f %9.5f %9.5f %9.5f"%(unpolar_miev[0],unpolar_miev[1],unpolar_miev[2],unpolar_miev[3]))
print("miepython %9.5f %9.5f %9.5f %9.5f"%(unpolar[0],unpolar[1],unpolar[2],unpolar[3]))
print("ratio %9.5f %9.5f %9.5f %9.5f"%(ratio[0],ratio[1],ratio[2],ratio[3]))
MIEV0 Test Case 14: m=1.500-1.000j, Mie size parameter = 1.000
0.0° 30.0° 60.0° 90.0°
MIEV0 0.37745 0.31321 0.19214 0.12066
miepython 0.37745 0.31321 0.19214 0.12066
ratio 1.00000 1.00000 1.00000 1.00000
Wiscombe’s Test Case 10¶
[9]:
"""
MIEV0 Test Case 10: Refractive index: real 1.330 imag -1.000E-05, Mie size parameter = 100.000
Angle Cosine S-sub-1 S-sub-2 Intensity Deg of Polzn
0.00 1.000000 5.25330E+03 -1.24319E+02 5.25330E+03 -1.24319E+02 2.76126E+07 0.0000
30.00 0.866025 -5.53457E+01 -2.97188E+01 -8.46720E+01 -1.99947E+01 5.75775E+03 0.3146
60.00 0.500000 1.71049E+01 -1.52010E+01 3.31076E+01 -2.70979E+00 8.13553E+02 0.3563
90.00 0.000000 -3.65576E+00 8.76986E+00 -6.55051E+00 -4.67537E+00 7.75217E+01 -0.1645
"""
x=100.0
m=1.33-1e-5j
mu=np.cos(np.linspace(0,90,4) * np.pi/180)
qext, qsca, qback, g = miepython.mie(m,x)
albedo = qsca/qext
unpolar = miepython.i_unpolarized(m,x,mu,'wiscombe')
unpolar_miev = np.array([2.76126E+07,5.75775E+03,8.13553E+02,7.75217E+01])
ratio = unpolar_miev/unpolar
print("MIEV0 Test Case 10: m=1.330-0.00001j, Mie size parameter = 100.000")
print()
print(" %9.1f°%9.1f°%9.1f°%9.1f°"%(0,30,60,90))
print("MIEV0 %9.0f %9.0f %9.0f %9.0f"%(unpolar_miev[0],unpolar_miev[1],unpolar_miev[2],unpolar_miev[3]))
print("miepython %9.0f %9.0f %9.0f %9.0f"%(unpolar[0],unpolar[1],unpolar[2],unpolar[3]))
print("ratio %9.5f %9.5f %9.5f %9.5f"%(ratio[0],ratio[1],ratio[2],ratio[3]))
MIEV0 Test Case 10: m=1.330-0.00001j, Mie size parameter = 100.000
0.0° 30.0° 60.0° 90.0°
MIEV0 27612600 5758 814 78
miepython 27612635 5758 814 78
ratio 1.00000 1.00000 1.00000 1.00000
Wiscombe’s Test Case 7¶
[10]:
"""
MIEV0 Test Case 7: Refractive index: real 0.750 imag 0.000E+00, Mie size parameter = 10.000
Angle Cosine S-sub-1 S-sub-2 Intensity Deg of Polzn
0.00 1.000000 5.58066E+01 -9.75810E+00 5.58066E+01 -9.75810E+00 3.20960E+03 0.0000
30.00 0.866025 -7.67288E+00 1.08732E+01 -1.09292E+01 9.62967E+00 1.94639E+02 0.0901
60.00 0.500000 3.58789E+00 -1.75618E+00 3.42741E+00 8.08269E-02 1.38554E+01 -0.1517
90.00 0.000000 -1.78590E+00 -5.23283E-02 -5.14875E-01 -7.02729E-01 1.97556E+00 -0.6158
"""
x=10.0
m=0.75
mu=np.cos(np.linspace(0,90,4) * np.pi/180)
qext, qsca, qback, g = miepython.mie(m,x)
albedo = qsca/qext
unpolar = miepython.i_unpolarized(m,x,mu,'wiscombe')
unpolar_miev = np.array([3.20960E+03,1.94639E+02,1.38554E+01,1.97556E+00])
ratio = unpolar_miev/unpolar
print("MIEV0 Test Case 7: m=0.75, Mie size parameter = 10.000")
print()
print(" %9.1f°%9.1f°%9.1f°%9.1f°"%(0,30,60,90))
print("MIEV0 %9.2f %9.2f %9.2f %9.2f"%(unpolar_miev[0],unpolar_miev[1],unpolar_miev[2],unpolar_miev[3]))
print("miepython %9.2f %9.2f %9.2f %9.2f"%(unpolar[0],unpolar[1],unpolar[2],unpolar[3]))
print("ratio %9.5f %9.5f %9.5f %9.5f"%(ratio[0],ratio[1],ratio[2],ratio[3]))
MIEV0 Test Case 7: m=0.75, Mie size parameter = 10.000
0.0° 30.0° 60.0° 90.0°
MIEV0 3209.60 194.64 13.86 1.98
miepython 3209.60 194.64 13.86 1.98
ratio 1.00000 1.00000 1.00000 1.00000
Comparison to Bohren & Huffmans’s Mie Program¶
Bohren & Huffman normalizes as
Bohren & Huffmans’s Test Case 14¶
[11]:
"""
BHMie Test Case 14, Refractive index = 1.5000-1.0000j, Size parameter = 1.0000
Angle Cosine S1 S2
0.00 1.0000 -8.38663e-01 -8.64763e-01 -8.38663e-01 -8.64763e-01
0.52 0.8660 -8.19225e-01 -8.61719e-01 -7.21779e-01 -7.27856e-01
1.05 0.5000 -7.68157e-01 -8.53697e-01 -4.19454e-01 -3.72965e-01
1.57 0.0000 -7.03034e-01 -8.43425e-01 -4.44461e-02 6.94424e-02
"""
x=1.0
m=1.5-1j
mu=np.cos(np.linspace(0,90,4) * np.pi/180)
qext, qsca, qback, g = miepython.mie(m,x)
unpolar = miepython.i_unpolarized(m,x,mu,norm='bohren')
s1_bh = np.empty(4,dtype=complex)
s1_bh[0] = -8.38663e-01 - 8.64763e-01*1j
s1_bh[1] = -8.19225e-01 - 8.61719e-01*1j
s1_bh[2] = -7.68157e-01 - 8.53697e-01*1j
s1_bh[3] = -7.03034e-01 - 8.43425e-01*1j
s2_bh = np.empty(4,dtype=complex)
s2_bh[0] = -8.38663e-01 - 8.64763e-01*1j
s2_bh[1] = -7.21779e-01 - 7.27856e-01*1j
s2_bh[2] = -4.19454e-01 - 3.72965e-01*1j
s2_bh[3] = -4.44461e-02 + 6.94424e-02*1j
# BHMie seems to normalize their intensities to 4 * pi * x**2 * Qsca
unpolar_bh = (abs(s1_bh)**2+abs(s2_bh)**2)/2
ratio = unpolar_bh/unpolar
print("BHMie Test Case 14: m=1.5000-1.0000j, Size parameter = 1.0000")
print()
print(" %9.1f°%9.1f°%9.1f°%9.1f°"%(0,30,60,90))
print("BHMIE %9.5f %9.5f %9.5f %9.5f"%(unpolar_bh[0],unpolar_bh[1],unpolar_bh[2],unpolar_bh[3]))
print("miepython %9.5f %9.5f %9.5f %9.5f"%(unpolar[0],unpolar[1],unpolar[2],unpolar[3]))
print("ratio %9.5f %9.5f %9.5f %9.5f"%(ratio[0],ratio[1],ratio[2],ratio[3]))
print()
print("Note that this test is identical to MIEV0 Test Case 14 above.")
print()
print("Wiscombe's code is much more robust than Bohren's so I attribute errors all to Bohren")
BHMie Test Case 14: m=1.5000-1.0000j, Size parameter = 1.0000
0.0° 30.0° 60.0° 90.0°
BHMIE 1.45117 1.23221 0.81695 0.60621
miepython 1.50978 1.25285 0.76857 0.48265
ratio 0.96118 0.98353 1.06296 1.25600
Note that this test is identical to MIEV0 Test Case 14 above.
Wiscombe's code is much more robust than Bohren's so I attribute errors all to Bohren
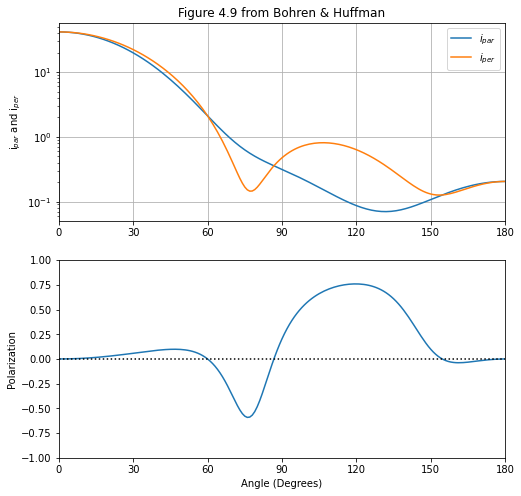
Bohren & Huffman, water droplets¶
Tiny water droplet (0.26 microns) in clouds has pretty strong forward scattering! A graph of this is figure 4.9 in Bohren and Huffman’s Absorption and Scattering of Light by Small Particles.
A bizarre scaling factor of \(1/4\) is needed to make the miepython results match those in the figure 4.9.
[12]:
x=3
m=1.33-1e-8j
theta = np.linspace(0,180,181)
mu = np.cos(theta*np.pi/180)
scaling_factor = 1/4
iper = scaling_factor*miepython.i_per(m,x,mu,'bohren')
ipar = scaling_factor*miepython.i_par(m,x,mu,'bohren')
P = (iper-ipar)/(iper+ipar)
plt.subplots(2,1,figsize=(8,8))
plt.subplot(2,1,1)
plt.semilogy(theta,ipar,label='$i_{par}$')
plt.semilogy(theta,iper,label='$i_{per}$')
plt.xlim(0,180)
plt.xticks(range(0,181,30))
plt.ylabel('i$_{par}$ and i$_{per}$')
plt.legend()
plt.grid(True)
plt.title('Figure 4.9 from Bohren & Huffman')
plt.subplot(2,1,2)
plt.plot(theta,P)
plt.ylim(-1,1)
plt.xticks(range(0,181,30))
plt.xlim(0,180)
plt.ylabel('Polarization')
plt.plot([0,180],[0,0],':k')
plt.xlabel('Angle (Degrees)')
plt.show()
Comparison to PyMieScatt¶
This package evidently uses Wiscombe’s normalization.
import PyMieScatt
print(PyMieScatt.MieS1S2(1.5,0.02,0.75))
print(miepython.mie_S1_S2(1.5,0.02,0.75,norm='wiscombe'))
produces:
::
((3.690888119953865e-12-2.353141061130335e-06j), (2.7681660899653986e-12-1.7648752402921955e-06j)) ((3.690978962673683e-12+2.3531410334006517e-06j), (2.7682310595192753e-12+1.7648752220853264e-06j))
[ ]: